Executive Summary 2019 Report
Project rationale and overall objectives of the project
The ULTRA-DD project’s goal is to deliver new tools and resources to speed up the development of truly innovative medicines, especially in the areas of autoimmune and inflammatory diseases, where new treatments are urgently needed. Through the partner Structural Genomics Consortium (SGC), ULTRA-DD has strong ties with similar initiatives elsewhere in the world, and this, coupled with the project’s strong open science policy ensures that the tools, resources, and knowledge generated by the project benefit the entire scientific community and enable new drug discovery and development projects within the industrial biomedical sector.
Overall deliverables of the project
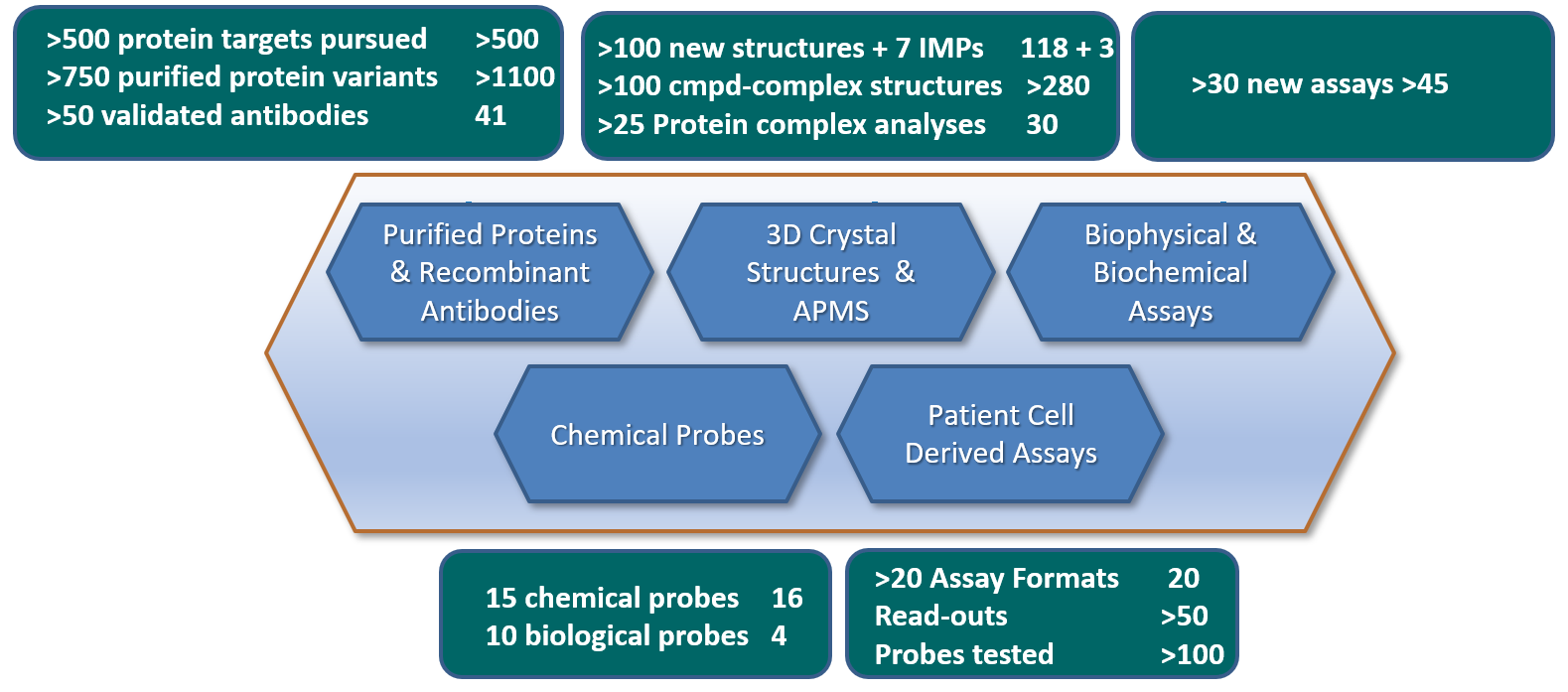
Figure 1. Overview of all main deliverables for ULTRA-DD during the period 2015-2020. Deliverables status as of 28th of February 2019 (end P4) is annotated in the right hand of each deliverables box.
Summary of progress versus plan since last period
All Work-Packages (WPs) have met their milestones and deliverables during period 4. Further, all WPs are on track towards the final milestones and deliverables due at the end of the project in reporting period 5 (M49-60). There have been no deviations from plan other than that certain WPs have over-delivered or reached certain milestones and deliverables ahead of plan.
Significant achievements since last report
WP1 - Target Prioritization
We have achieved all goals for the reporting period. Target priority lists have been updated every 6 months for: (i) chemical probe targets; (ii) structural targets and (iii) antigens for antibody production. The lists have been updated on 1st November 2018 and on 28th February 2019, alongside the 7th and 8th TPN teleconference meeting. The list currently comprises 1,020 protein targets, including 111 targets delivered from Pi (Priority index, our in-house genetics-led tool).
WP2 - Protein and Antibody Production
We have over-achieved the goals also for year 4. We are continuing to exceed our deliverables for this work package with 5915 protein constructs generated by the end of P4, resulting in a total (P1 - P4) of 766 purified soluble proteins, 309 purified integral membrane proteins (IMPs) and 124 antigens. To date, antibodies against 41 targets have passed validation (including IP-MS) and thus been shown to bind their full-length endogenous target from a cell lysate (eight of these targets in P4). An increase in the use of mammalian expression has also occurred, with focused efforts on IMPs and other difficult to express targets.
WP3 - Cell Assays
All milestones and deliverables have been met. The focus of the current reporting period has been on the YEATS epigenetic reader family (MLLT1, YEATS2, MLLT3, YEATS4) for which more than 200,000 data points have been generated. With three new active chemical probe projects initiated for the new NUDIX family (in particular NUDT5, NUDT7, and NUDT21) a further 59,000 data points have been generated. In total, in vitro screening has been performed on 10 targets.
WP4 - Protein Structures and Proteomics
We have met all milestones and deliverables. We solved 35 structures (4 novel and 31 follow-up) of soluble ULTRA-DD targets from 14 targets. We have completed and deposited the structure of the integral membrane protein HCN4, and two additional novel IMP structures are in refinement. We have undertaken 5 crystallographic fragment screening campaigns (D4.4), and deposited fragment screens on 4 targets (NUTD5, NUDT7, PARP14, DCP2B), comprising a total of 131 protein-fragment structures. We successfully analysed 7 protein complexes by mass spectrometry (MS) during the year (30 in total).
WP5 - Chemical Probes
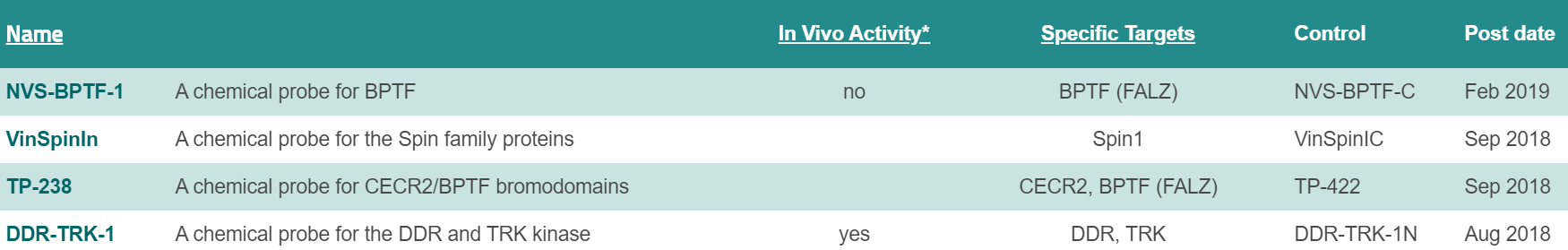
All milestones have been met or exceeded in year 4, culminating in the approval of 4 new chemical probes (16 approved probes in total for P1-P4). As previously reported, our scientific focus has shifted from bromodomain and lysine demethylase targets to the YEATS, readers of acetyl and crotonylated lysine residues. Vinspinin and iMLLT were two first in class chemical probes in the past year for the spindlin methyl lysine readers and the YEATS acyl lysine readers. Work on fragment, high-throughput and cellular screening for NUDIX proteins has continued, and we expect to nominate our first NUDIX probe within the next six months. The NUDT7 probe, if meeting all quality criteria, will if approved, be the first irreversible, covalent chemical probe from ULTRA-DD. We have also broadened our target portfolio to include E3 ligases of the ubiquitin system, including developing new informatics and analysis tools (see WP7 below).
WP6 - Patient-Derived Cell Assays
All milestones and deliverables have been met. During the reporting period, KI has developed two new patient derived cell assays: i) IBD whole blood assay MDP, analysing effects on release of pro-inflammatory cytokines (109 chemical probes; 23 patients and 6 healthy donors) and ii) skin fibroblast assay analysing effects on live cell imaging parameters in a scratch assay, as well as biomarkers (83 chemical probes; 5 Systemic sclerosis patients and 5 healthy donors). UOXF has developed new assays for Dupuytren’s Disease: i) Viability/proliferation/confluency assay (75 chemical probes, 5 patients); ii) Periostin secretion assay (54 chemical probes, 4 patients), and iii) pro-fibrotic PCR genes expression assay (21 chemical probes, 5 patients). In Spondyloarthritis, we have set up two new assays: i) Proliferation/viability assay and ii) M1 Macrophage cytokine secretion assay, analysing IL-12 and IL-23 release (54 chemical probes, 12 patients). Data and experimental protocols for 10 assays have to date been published (pre-publication) on the ULTRA-DD webpage https://ultra-dd.org/tissue-platforms.
WP7 - Informatics
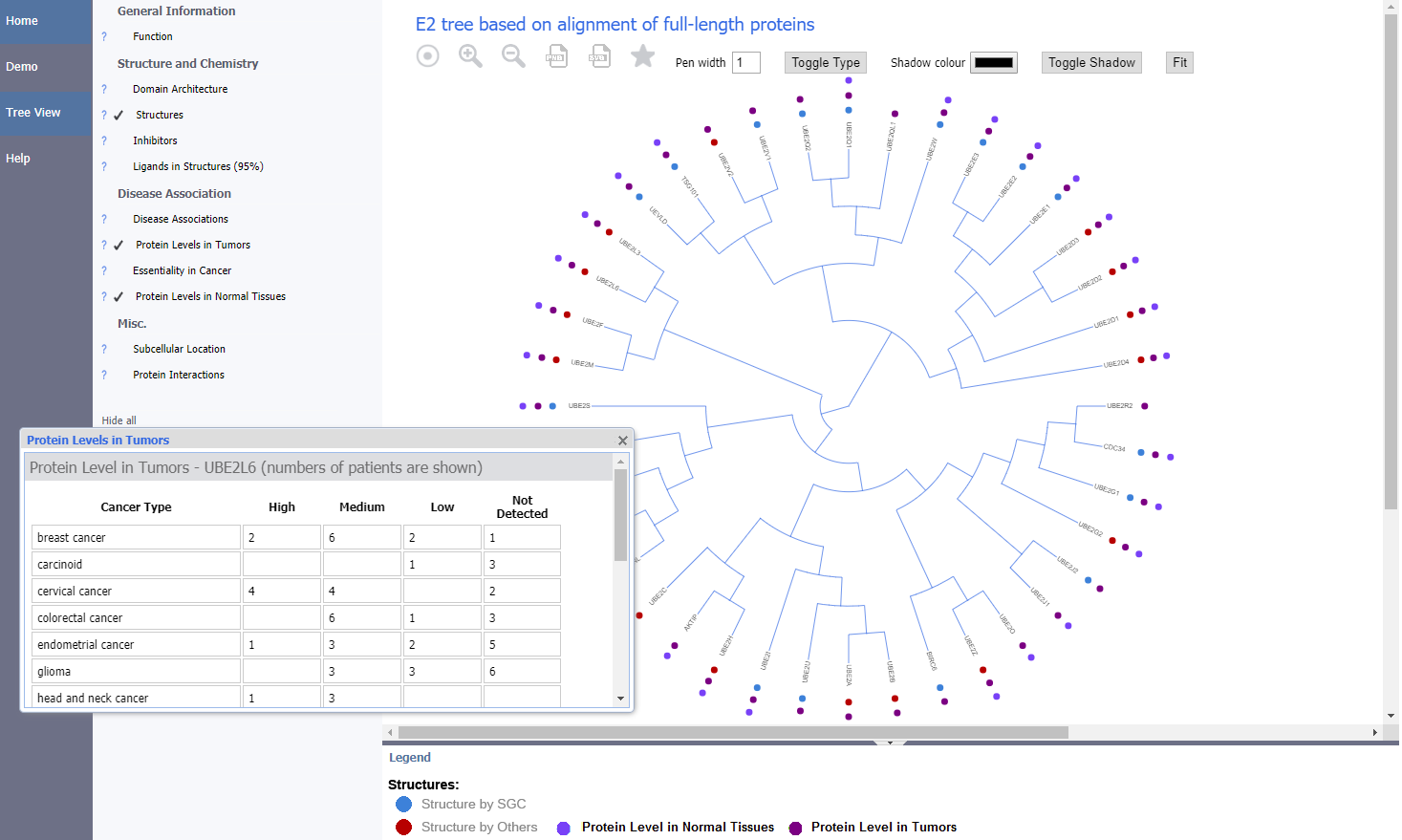
All milestones and deliverables were met. Working in collaboration with SGC Toronto we have developed a new web portal, called UbiHub - https://ubihub.thesgc.org, which allows visitors to explore information available for protein families involved in ubiquitin mediated pathways by overlaying annotations onto phylogenetic trees covering these families. The new portal re-uses and extends the technology developed for the previously reported ChromoHub tool to include new types of annotation and phylogenetic tree rendering styles (https://chromohub.thesgc.org)
WP8 - Outreach and Network
All milestones and deliverables were met. ULTRA-DD chemical probes have been distributed directly by us to 48 international research groups and over 200 samples (including entire sets) were distributed through our chemical vendor partners. A major new project funded by several patient groups around ALS (Amyotrophic lateral sclerosis, also known as Motor Neuron Disease) has been started to generate and characterise reproducible antibodies. Further funding from the disease foundations, FOP Friends and The Brain Tumour Charity, to pursue small molecule design for ALK2 has been received. In addition, through the linked effort at Oxford for inflammation TEPs (target enabling packages), further leveraging of funding and outputs have been achieved (TEP project is funded by the Wellcome Trust).
WP9 - Management
All milestones and deliverables were met. Overall, the management of the project continues to work well with the established governance structure. The SGC company continues to monitor and coordinate the project finances in close cooperation with the academic institutions. Research activities are running according to budget. The annual ULTRA-DD conference was hosted by University of Oxford in April 2018 with a mix of presentations from collaborators and our junior faculty and students and the annual conference for P4 will be held at ETHZ on the 8th of May 2019.



