Executive Summary 2020 Report
Project rationale and overall objectives of the project
The ULTRA-DD project’s goal is to deliver new tools and resources to speed up the development of truly innovative medicines, especially in the areas of autoimmune and inflammatory diseases, where new treatments are urgently needed. Through the partner Structural Genomics Consortium (SGC), ULTRA-DD has strong ties with similar initiatives elsewhere in the world, and this, coupled with the project’s strong open science policy ensures that the tools, resources, and knowledge generated by the project benefit the entire scientific community and enable new drug discovery and development projects within the industrial biomedical sector.
Overall deliverables of the project
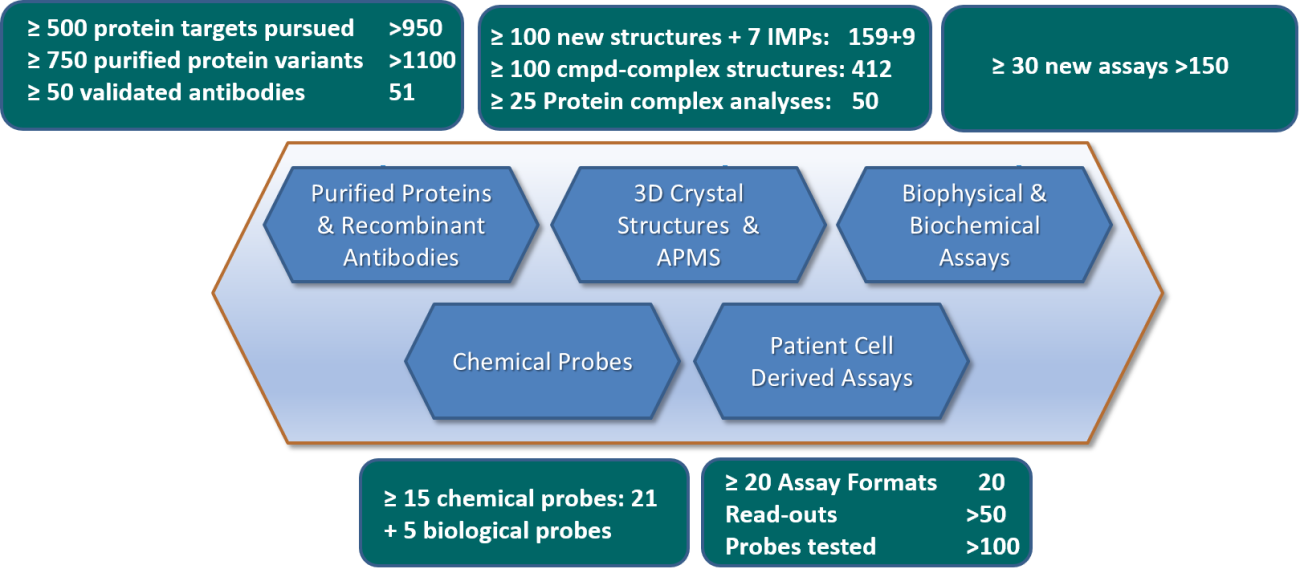
Figure 1. Overview of all main deliverables for ULTRA-DD during the period 2015-2020. Deliverables status as of 28th of February 2020 (at the end of the project following reporting period 5) is annotated in the right hand of each deliverables box.
Summary of progress versus plan since last period
All Work-Packages (WPs) have met their milestones and deliverables also during period 5. Further, all WPs reached their final milestones and deliverables at the end of the project in reporting period 5 (M49-60). There have been no deviations from plan throughout reporting periods 1-5, other than that certain WPs have over-delivered or reached certain milestones and deliverables ahead of plan.
Significant achievements since last report
WP1: All milestones and deliverables have been met or exceeded. The final target list covers (i) chemical probe targets; (ii) structural targets and (iii) antigens for antibody production. In particular the list includes 167 targets with high or medium priority, that (i) are nominated by TPN partners consolidating evidence relevant to inflammatory disease; and/or (ii) are evidenced from our in-house genetics-led prioritisation tool (Priority Index, Pi) applied to 30 immune-related traits.
WP2: All milestones and deliverables have been met or exceeded. Our BacMam expression platform has continued to enable the expression of new targets and protein complexes over the past year. We improved steps in the process by changing our insect transfection reagent and also acquired a new cell line from Thermo Fisher (Expi293 GnTI) that grows faster and more consistently. This cell line has enabled the production of new membrane proteins and structures. Antibodies for 10 targets have passed IP-MS validation and shown to bind their full-length endogenous target from a cell lysate. The generated biological probe for Interleukin-2 (IL2) was shown to neutralize IL2 in a functional assay.
WP3: All milestones and deliverables have been met or exceeded. During P5, efforts were focused on the YEATS (MLLT1, YEATS2, MLLT3, YEATS4) and NUDIX (NUDT5, NUDT7, NUDT21) families for which several new cell assays have been generated. Three new chemical probe candidates were identified for YEATS and cell-active compounds were obtained for both NUDT5 and NUDT7. Additional HTS assays for several ubiquitin-related targets have been established.
WP4: All milestones and deliverables have been met or exceeded. We solved 15 structures (4 novel and 11 follow-up) of soluble ULTRA-DD targets from 7 targets. We have undertaken 3 crystallographic fragment screening campaigns (ACVR1A, DHTKD1A, INPP5D), and deposited fragment screens of 5 targets (ATAD2A, FALZA, NUDT22A, TNCA, YEATS4A). The main objective in the fifth period has been to complete our ongoing complex analysis and prepare the obtained results for publication. Specifically, we have completed our analysis of (i) kinase complexes, (ii) the study on the modular organization of the prefoldin protein family and (iii) the quantitative analysis of TNFalpha receptor complexes. Moreover, we identified previously unknown neosubstrate for Cereblon induced by Lenalidomide and Pomalidomide.
WP5: All milestones and deliverables have been met or exceeded. We have delivered 21 chemical probes in total, six more than the official goal of 15. The last year has seen a continued focus on the Nudix protein domains. In addition, major screening efforts were directed at the KELCH domain containing proteins, KLHDC2, KLHL20 and KLHL3. The KELCH domains are E3 ligase substrate recognition domains and future chemical probes for them will be useful for both studying their function and potentially as new E3 domains for proteolysis targeted chimera (PROTAC) development.
WP6: All milestones and deliverables have been met. The main focus for our activities during P5 has been on developing advanced cell assays for the study of epithelial cell biology in IBD. These include development of colon epithelial organoids and 2D epithelial cell cultures, as well as studies on freshly obtained mucosal biopsies from patients with IBD. Target validation of potential novel intervention points for modulation of autoimmune diseases, focusing on SLE, has been productive. We also developed a new wound healing assay for Dupuytren’s Disease (hand fibrosis).
WP7: All milestones and deliverables were met. In addition to ensuring chemical probes, antibodies and structure deliverables are deposited in suitable repositories and made available via the ULTRA-DD website, we have continued to ensure that new tissue platform assay data and metadata are made publically available (https://ultra-dd.org/tissue-platforms/cell-assay-datasets). Work has commenced in a collaboration with the FAIRplus IMI project to identify ways to ensure that these data are made available in a FAIR-compliant manner in the near future.
WP8: All milestones and deliverables were met. Numerous academic collaborations have been initiated during P5. These include clinical proteomics, transcriptomics and spatial transcriptomics studies in IBD ULTRA-DD 6 with the Swedish national research infrastructure at Science for Life Laboratory, two new collaborating hospitals for patient material access (Karolinska University Hospital South and Ersta Hospital); and many academic collaborations, e.g. with the Goethe University in Frankfurt and the University of North Carolina for chemical probes targeting kinases.
The most advanced translational medical studies, which we highlighted in the P4 report, arising in part from ULTRA-DD outputs and staff, continues according to plan (the M4K Pharma and IBD clinical translation). In addition, our tissue platform team and other scientists at UOXF received additional funding for translational studies in e.g. fibrosis, from Evotec through their Lab282 program https://www.lab282.org/about/funded-projects.
WP9: All milestones and deliverables were met. The established governance structure for project management has worked well also during the final year of the project. The SGC company has continued to monitor and coordinate the project finances in close cooperation with the academic partners. The annual ULTRA-DD conference (for P4) was hosted by ETH on 8th May 2019 and featured project updates by junior faculty as well as presentations from local experts on the latest technology developments in mass spectrometry and patient profiling. The final ULTRA-DD conference was held virtually due to the ongoing SARS-CoV-2 pandemic on March 24th 2020.
Scientific and technical results/foregrounds of the project
For the entire project period (years 1 – 5), the ULTRA-DD scientists, EFPIA partners and collaborators have generated the following main foreground (see also Figure 1 above):
- >7000 expression constructs for >900 target proteins
- SGC plasmid collection at Addgene
- Gräslund lab plasmid collection at Addgene
- >1100 purified protein samples
- >50 high quality and renewable antibodies, of which five (5) declared as biological probes
- ULTRA-DD antibodies
- ImmunoPrecise
- 168 novel protein structures, of which nine (9) were from integral membrane proteins
- ULTRA-DD structures
- Analyses of 50 protein complexes by advanced mass spectrometry
- Publication
- Raw data
- Filtered interaction data
- 190 new cell assays, of which 20 were derived from patient material (blood, biopsies), some of which were included in the following publications:
- NanoBRET for CDK4/6
- FRAP for NSD3
- AlphaScreen, chemoproteomics & NanoBRET for SPIN1
- CETSA for NUDT7
- Chemoproteomics for PRMTs & EED
- AlphaScreen TR-FRET, NanoBRET & CETSA for MLLT1/3
- Chemoproteomics for RIPK2



