Executive Summary 2018 Report
Project rationale and overall objectives of the project
The ULTRA-DD project’s goal is to deliver new tools and resources to speed up the development of truly innovative medicines, especially in the areas of autoimmune and inflammatory diseases, where new treatments are urgently needed. Through the partner Structural Genomics Consortium (SGC), ULTRA-DD has strong ties with similar initiatives elsewhere in the world, and this, coupled with the project’s strong open access policy ensures that the tools, resources, and knowledge generated by the project benefit the entire scientific community and enable new drug discovery and development projects within the industrial biomedical sector.
Overall deliverables of the project
A summary of all deliverables for ULTRA-DD is found in Figure 1 below
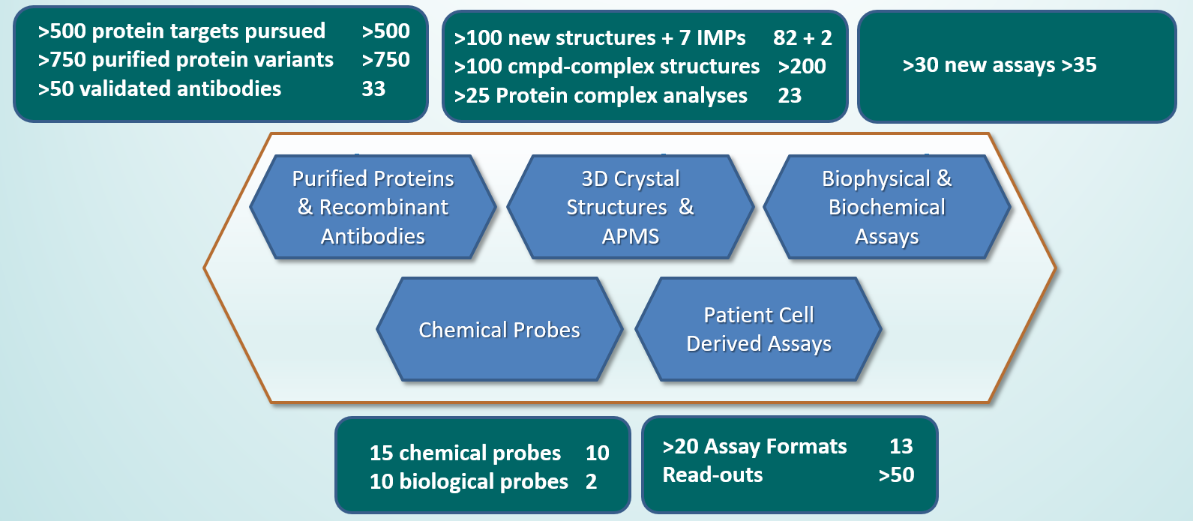
Figure 1. Overview of all deliverables for ULTRA-DD during the period 2015-2020. Deliverables status as of 28th of February 2018 is annotated in the right hand of each deliverables box.
Summary of progress versus plan since last period
All Work-Packages (WPs) have met their milestones and deliverables during the period M15-M36. Further, all WPs are on track towards upcoming major milestones and deliverables due in reporting period 4 (M37-48). There have been no deviations from plan other than that certain WPs have over-delivered or reached certain milestones ahead of plan.
Significant achievements since last report
WP1 – Target Prioritization
We have achieved all goals for the reporting period. Target lists have been updated continuously with major revisions on 15th March 2017 and on 27th March 2018, alongside the 5th TPN teleconference meeting plus a special meeting (6th TPN) seeking the opportunities for validation using Janssen in-house L1000 drug/compound expression data. The list currently comprises 909 protein targets.
WP2 – Protein and Antibody Production
We have over-achieved the goals also for year 3. We have by end of P3 generated in total 4593 protein expression constructs during years 1-3. This has enabled the delivery of 701 purified soluble proteins and 194 integral membrane proteins (IMP) samples for various analyses in WP3, 4 and 5 (e.g. assays and screening). In addition, 104 antigens were generated within this work package, which to date has led to the production of 33 high quality antibodies. We have also tested and optimized new methods for expression and characterization of IMPs as well as mammalian expression for antigen production.
WP3 – Cell Assays
All milestones and deliverables were met. Screening efforts were mainly directed at epigenetic targets including the YEATS domain epigenetic reader protein family (MLLT1, YEATS2, MLLT3, YEATS4) as well as the Kme reader SPIN1. In total in vitro screening has been performed on 22 targets and approximately 120,000 data points have been generated.
WP4 – Protein Structures and Proteomics
We have met all milestones and deliverables. We have 20 novel and follow-up structures of soluble ULTRA-DD targets, including 3 novel structures and 17 follow-up structures from 5 targets. We have completed and deposited fragment screens on 10 targets (PHIP, BAZ2B, ATAD2, FALZ, SP100, BRD1, JMJD2D, NUDT22, JARID1B, JMJD1B) and a further six screens (NUDT4, NUDT7, NUDT21, DCP2, PARP14A and MACROD1) have been completed and are ready for deposition (WP4.4). We have completed and deposited the structure of the integral membrane protein ABC transporter, ABCB8, with ADP. ETH Zürich successfully analyzed 10 protein complexes during the year (23 in total). In addition, the ETH team has identified a high-resolution interaction map of activated TNF-alpha receptor complexes using a ligand binding approach.
WP5 – Chemical Probes
All milestones have been met or exceeded in year 3, culminating in the approval of 4 new chemical probes. In the epigenetic space, the scientific focus has shifted from the previous bromodomain and lysine demethylase targets to the YEATS, readers of acetyl and crotonylated lysine residues. In addition, we have made good progress in fragment, high-throughput and cellular screening for Nudix proteins.
WP6 – Patient-Derived Cell Assays
We met all milestones and deliverables but one and exceeded some of the goals. We have acquired cells or tissues from Ankylosing Spondylitis and Dupuytren’s Disease, Myositis, Lupus and Systemic sclerosis patients, and established 13 assay formats which have been used for probe testing. Access to FOP patient material has been achieved through collaboration with Alex Bullock at SGC Oxford. Further, new dedicated collaborative funding for studies in inflammatory bowel diseases was secured by July 1 2018. An agreement with the Karolinska University Hospital has been made to source patient material from Sjögren’s syndrome patients, with start second half of 2018.
WP7 – Informatics
All milestones and deliverables were met. The team has implemented new data-capture filters within the Scarab database to capture new assays developed in the last year at Karolinska Institutet and University of Oxford. We have also conducted a major redesign of the ULTRA-DD web-site to make it easier to navigate and view our deliverables thus far. With regards to data dissemination we have continued to work to improve the ULTRA-DD web-site and work with SGC Toronto on the maintenance and development of the ChromoHub tool. We have also visited the ChEMBL development team at the EBI to discuss the deposition of both SGC and ULTRA-DD chemical probe datasets into the ChEMBL database for which we hope the first datasets will be deposited by July 2018.
WP8 – Outreach and Network
All milestones and deliverables were met. Our work to attract collaborations with Disease Foundations and Patient Organizations has expanded with a larger collaborative effort with three ALS patient organizations. We are happy to report that a new research programme aiming to quantify and model economic impact driven by open science has been put in place in P3 by two of the ULTRA-DD Principal Investigators and funded by the Oxford Martin School. ULTRA-DD chemical probes have been distributed directly by us to at least 20 international research groups and through our chemical vendor partners, additional ~200 samples were distributed, although we cannot with certainty know from how many individual organizations. We have during P3 also initiated a collaboration with the company Modiquest for distribution of open access antibodies.
WP9 – Management
All milestones and deliverables were met. Overall, the management of the project is working well, with established informal and formal channels for governance and interactions. Project finances are coordinated from University of Toronto, and research activities are running according to budget. A team of principal investigators, scientists and PhD students from ULTRA-DD alongside EFPIA partners visited Brussels and the IMI in October 2017 for the half-way review. Overall, the feedback from the review committee was very positive.



